Example Files
Example scripts and data are included in the HiTRACE repository. For a new analysis, start with these example scripts and replace relevant values with yours:
-
[NEW] For further practice, we provide 12 more examples. They are previous 1D and 2D datasets annotated by our experts. Download Script & Data, and follow these instructions.
Workspace files are provided for comparison only, when you want to double check if your results are the same (especially for Step #4). They are not needed for trying out and starting fresh.
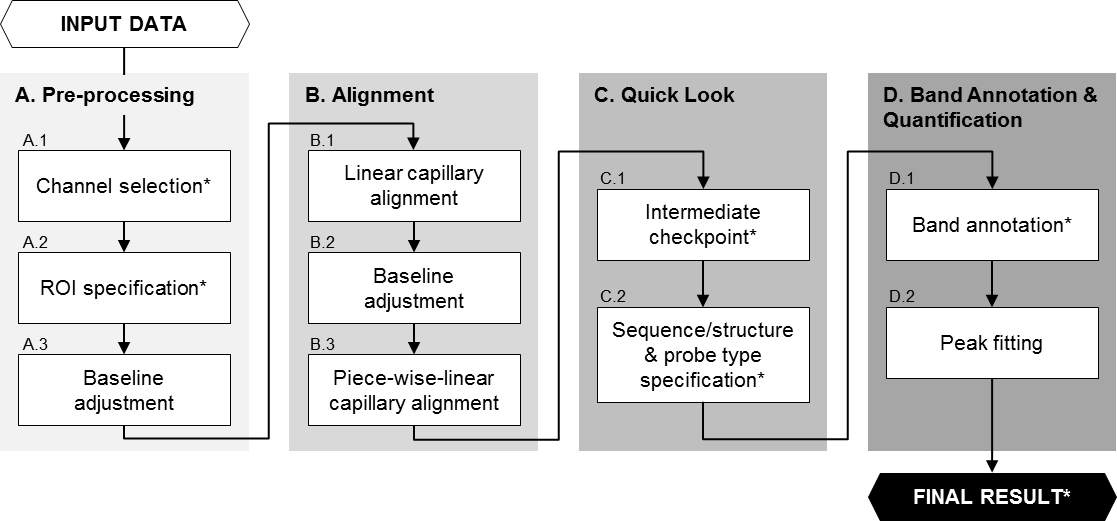
Analysis Pipeline
The complete HiTRACE analysis pipeline is composed of the following steps.
-
#0: Get Started
Checklist before kick off.
-
#1: Load and Preview Data
Read in data and provide a quick look.
-
#2: Further Alignment (optional)
Perform additional finer alignments.
-
#3: Parameter Input
Input experiment details and basic RNA construct information.
-
#4: Sequence Assignment
Annotate the position of each nucleotide position on the gel matching bands.
-
#5: Peak Fitting
Integrate band intensities into nucleotide peak readings.
-
#6: Data Corrections (for 1D data only)
Perform unsaturation, attenuation correction, background subtraction, and normalization for 1D data.
-
#7: Error Estimates (for 1D data only)
Evaluate data and estimate errors across replicates.
-
#8: Save your Work
Print well-annotated figures of your analysis for archiving.
-
#9: Output RDAT File
Write analyzed result to file in RDAT format, for use of Z-score and prediction.
-
#10: Calculate Z-score (for 2D data only)
Transfrom 2D chemical mapping data into Z-score for use as 2D pseudo-free energy bonus for RNAstructure.
-
#11: Predict Secondary Structure
Run structural prediction using RNAstructure based on chemical mapping reactivity data, with bootstrapping for helix-wise confidence score.
-
#12: Visualize Secondary Structure (optional)
Draw secondary structure using VARNA with nucleotides color-coded by reactivity, and mark difference between secondary structure models.
Bonus Graphic Tools
-
Color 2D Diagram
Make publication-ready diagram of secondary structures with nucleotides color-coded by reactivity.
-
Color 3D Model
Make publication-ready 3D models with nucleotides color-coded by reactivity.
Share your Data
-
Submit to RMDB
Share your chemical mapping results to RNA Mapping Database.