Step 2: Further Alignment (optional)
After Step #1, small differences in mobilities in mutants are still apparent (“waviness”). For 2D datasets, the shifts are actually due to differences in sequence between different lanes. We now are interested in fine-tuning this alignment.
We need to describe which profiles to align to each other. We should only let profiles that are of same nature to be aligned to each other. For example, lanes of the same modifier (e.g. DMS), or lanes of similar chemistry (e.g. 1M7 and NMIA, both SHAPE).
ddNTPs sequencing ladders should not be aligned to each other, or to modifer profiles, because they have strong bands in totally different places.
align_blocks = {1:8, 9:24, 25:40, 41:56, 57:58, 59:60, 61:62, 63:64};
d_align_before_more_alignment = d_align;
d_align_dp_fine = align_by_DP_fine(d_align_before_more_alignment, align_blocks);
The numberings in
align_blocksare based on column indices ofd_align; not to be confused withreorderpreviously. Simply speaking, thereordernumbering of ABI sequencer files (.ab1) is no longer relevant after Step #1!
The above code specified 8 groups for finer alignment in align_blocks. It treats nomod, DMS, CMCT, SHAPE, ddATP, ddTTP, ddCTP, and ddGTP separately from each other. It also copies the d_align into d_align_before_more_alignment as a back-up, in case align_by_DP_fine() yields unsatisfactory results.
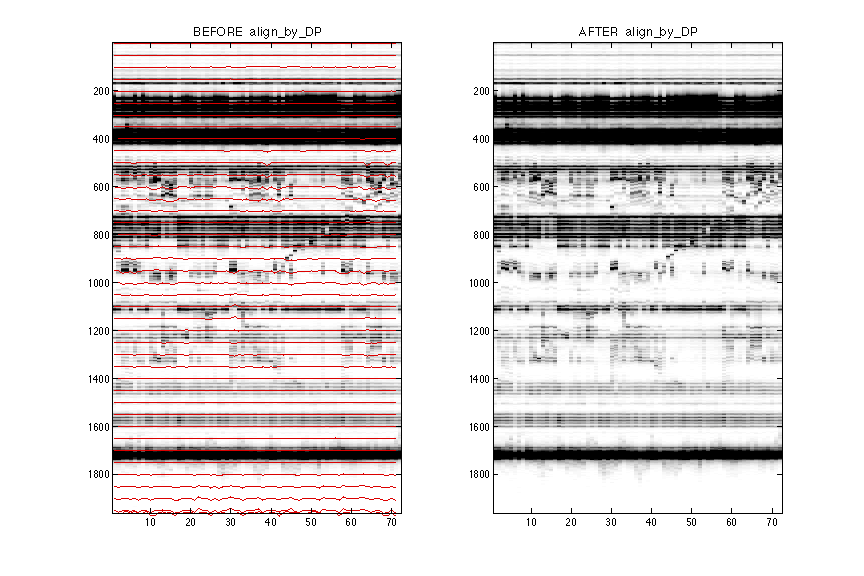
You’ll see a figure window showing the result side by side. It is obvious that after further alignment the “waviness” is minimized. The red lines are guidelines of alignment anchors. (In this case, due to multiple align_blocks, it is misplaced. Please ignore.)
It is important to check if the further alignment introduces artifacts. Look for bands that are shifted around too much and decide whether to accept it. You can get an idea on using all ddNTPs lanes as one
align_blocksand examine the obvious artifacts of forcing bands to align to each other.
There are other commands, e.g. align_by_DP() and align_by_DP_using_ref(), that performs alignment refinement in a different flavor. You may want to try those as well. You can also further-align on top of the result of align_by_DP_fine(). Make sure to backup your d_align.
Input arguments for
align_by_DP()andalign_by_DP_using_ref()may differ fromalign_by_DP_fine(). Usehelp()to double check.
To accept your further alignment, run:
d_align = d_align_dp_fine;
As an example of 2D data analysis, it is the same as 1D for this step.
align_blocks = {[1:55, 57:72]};
d_align_before_more_alignment = d_align;
d_align_dp_fine = align_by_DP_fine(d_align_before_more_alignment, align_blocks);
d_align = d_align_dp_fine;
This alignes all lanes (except 56) in 1 block. Lane 56 has bad data, thus is excluded here.