Examples of LIFFT runs
Ligand binding transitions
examples/eterna_fit_FMN_binding gives an example from the Eterna PNAS 2014 paper, fitting the binding affinity of flavin mononucleotide (FMN) to a couple of RNA sequences designed to present the FMN aptamer.
In MATLAB, change working directory to this directory. Then take a look at the example script RD031312_EteRNA_MinFMN_titrations8B_script_LIFFTtest.m. It reads in the
data from a workspace and then runs a loop over two data sets of FMN binding to different RNAs – run LIFFT on each.
open RD031312_EteRNA_MinFMN_titrations8B_script_LIFFTtest
RD031312_EteRNA_MinFMN_titrations8B_script_LIFFTtest
Example output, on second data set:
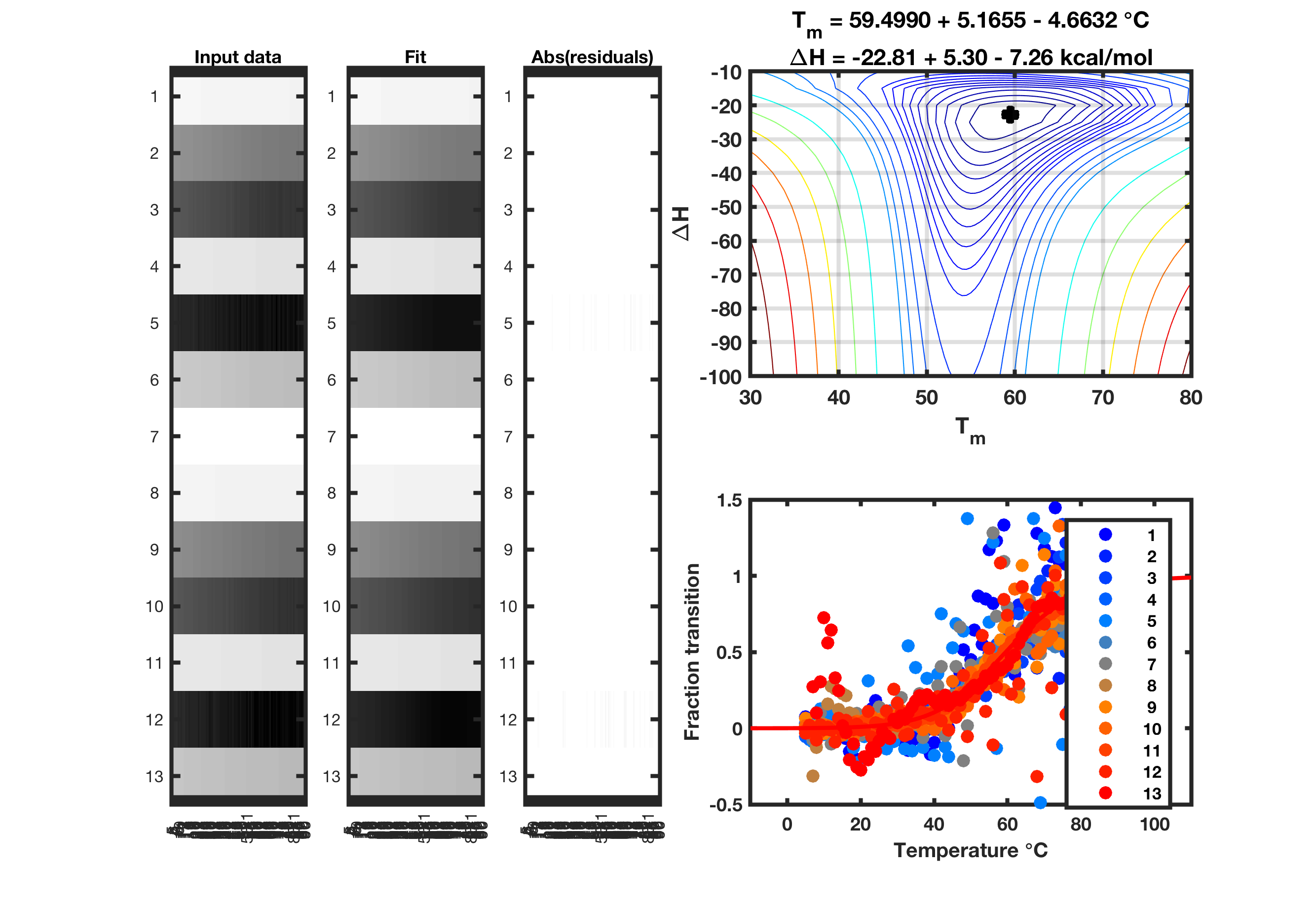
Temperature-dependent melts (chemical mapping)
examples/DMS_melt_AAAA gives an example of fitting to standardized dimethyl sulfate (DMS) data on an AAAA tetraloop hairpin, flanked by stable GAGUA reference hairpins.
Example output
Classic optical melts
Even classic optical melting experiments, read out by UV absorbance changes with temperature, can benefit from likelihood-based analysis. Shimadzu and Cary spectrophotometers often allow probing of the same nucleic acid sample at multiple concentrations. For monomeric unfolding, these should all report the same thermodynamic parameters, and so a global fit is possible. examples/optical_melt_AAAA gives an example of such a fit, including ‘sloping baselines’ and an update to take into account the lower point-to-point error in these experiments compared to chemical mapping.
Example output
- Previous: Documentation
- Next: Tips
Developed by Das lab, Leland Stanford Junior University.
Docs updated by Rhiju Das, March 2018.