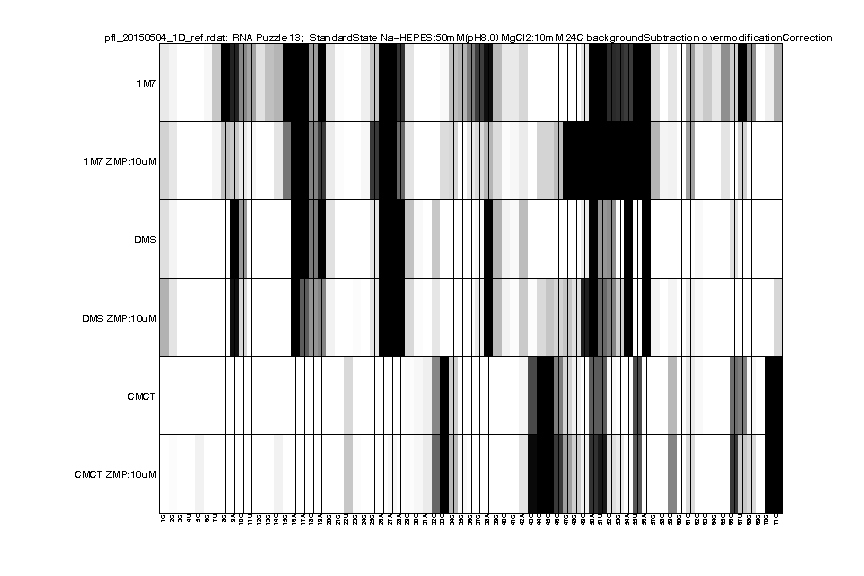
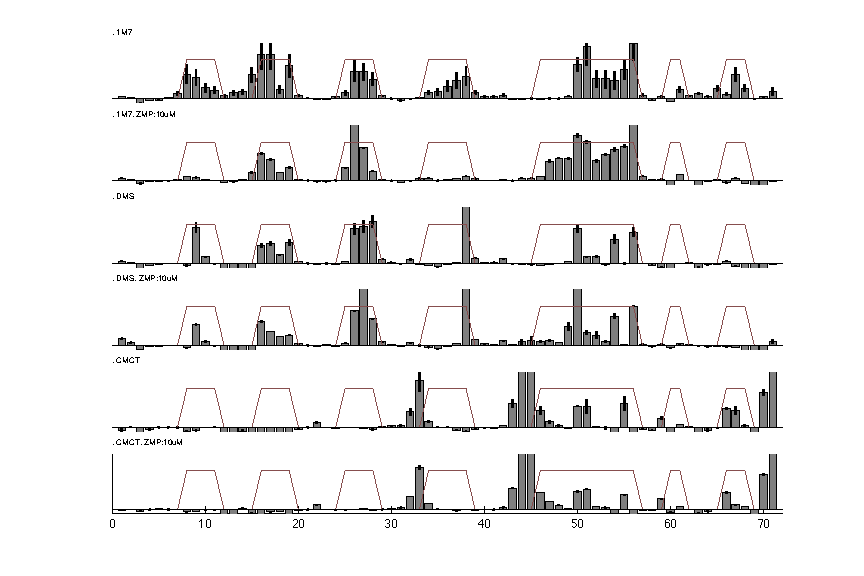
Step 9: Output RDAT File
This step requires proper setup of the package rdatkit. Please follow the installation guides there. Note that we only using the MATLAB components of rdatkit.
RDAT format is created for easy access and sharing data. It describes the annotation of electrophoresis traces such as these high-throughput experiments. It is the default format for the RNA mapping database and was designed to be an easily human-readable text file. In this tutorial, we will convert our analysis to RDAT file format as a nicely annotated and compact option.
To create a .rdat file, you need to define a few variables for the output – fortunately we have most of them already. For this case, you need to provide additionally a filename and the name of the RNA:
filename = 'pfl_1D.rdat';
name = 'RNA Puzzle 13';
Some overall annotations for the file. There as a small number of possible annotations here, like 'chemical', 'processing', etc. We are compiling a list of acceptable ones describing the specs. We also write a short comments for some notes:
comments = { ...
'Preliminary data, averaged over 2 replicates', ...
'RNA was heat to 90 C for 2 min and cool on ice for 2 min, then incubated at 37 C for 20 min at pH 8.0 with 10 mM MgCl2 to aid folding.', ...
'Additional sequences at 5'' and 3'' end not shown; two different flanking sequences were used and averaged.', ...
'Data are normalized based on GAGUA pentaloops in flanking sequences (not shown).', ...
'DMS, CMCT, and SHAPE normalized so that As, Us, or all 5 loop residues give mean reactivity of 1.0.', ...
'Note: ', ...
'Taken in early May 2015 for thirteenth RNA Puzzle community-wide modeling trial.', ...
};
annotations = { ...
'experimentType:StandardState', ...
'chemical:Na-HEPES:50mM(pH8.0)', 'chemical:MgCl2:10mM', ...
'temperature:24C', ...
'processing:backgroundSubtraction', 'processing:overmodificationCorrection', ...
};
As well as annotations specific for each profile:
data_annotations{1} = {'modifier:1M7'};
data_annotations{2} = {'modifier:1M7', 'chemical:ZMP:10uM'};
data_annotations{3} = {'modifier:DMS'};
data_annotations{4} = {'modifier:DMS', 'chemical:ZMP:10uM'};
data_annotations{5} = {'modifier:CMCT'};
data_annotations{6} = {'modifier:CMCT', 'chemical:ZMP:10uM'};
We use ‘uM’ instead of ‘μM’ to avoid character encoding issues.
Escape for ′ inside a string.
Here we decide to exclude the flanking sequence from the .rdat file publication. Thus, we need to trim our data and the associated variables. We figured out that in our seqpos_out, the region of interest is indexed from 25 to 95, spaning the length of 71 nucleotides. So that makes offset 0 now:
rdat_idx = 25:95;
rdat_seqpos = seqpos_out(rdat_idx); % 1:71
rdat_sequence = sequence(rdat_idx);
rdat_structure = structure(rdat_idx);
rdat_offset = 0;
rdat_d_final = [d_SHAPE_minus, d_SHAPE_plus, d_DMS_minus, d_DMS_plus, d_CMCT_minus, d_CMCT_plus];
rdat_da_final = [da_SHAPE_minus, da_SHAPE_plus, da_DMS_minus, da_DMS_plus, da_CMCT_minus, da_CMCT_plus];
rdat_d_final = rdat_d_final(rdat_idx, :);
rdat_da_final = rdat_da_final(rdat_idx, :);
Double check that the rows order is consistent with labels in
data_annotations.
Finally, we can create the RDAT file by:
output_workspace_to_rdat_file(filename, name, rdat_sequence, rdat_offset, rdat_seqpos, ...
rdat_d_final, rdat_structure, annotations, data_annotations, rdat_da_final, ...
[], [], [], comments);
We can read in the file for a visual check:
d_rdat = show_rdat(filename);
Once finished, the show_rdat() function returns a RDAT object:
| Variable | Type | Description |
|---|---|---|
d_rdat |
1x1 RDATFile | The RDAT file object. |
As an example of 2D data analysis, the commands are the same as 1D for this step. You only need to pay attention for the difference in data_annotations. Because it’s a Mutate-and-Map experiment, we need to label the mutations for each data lane. If you have the _keys.txt file from primerize design, you can use that directly:
construct_names = read_constructs('ZMP_2HP_keys.txt');
for j = 1:size(area_peak, 2);
data_annotations{j} = {['mutation:', strrep(strrep(strrep(construct_names{j}, 'T', 'U'), 'WU', 'WT'), 'Lib1-', '')]};
end;
If you don’t, generate it manually:
data_annotations{1} = 'mutation:WT';
for j = 2:size(area_peak, 2);
data_annotations{j} = {['mutation:', sequence(j - 1 - offset), num2str(j - 1), DNA2RNA(complement(sequence(j - 1 - offset)))]};
end;
Don’t forget to mark the bad lane:
data_annotations{56} = {'mutation:U55A', 'warning:badQuality'};
Also, since we did not label 'chemical' to each entry in data_annotations, we place it in annotations instead:
annotations = { ...
'experimentType:MutateAndMap', ...
'chemical:Na-HEPES:50mM(pH8.0)', 'chemical:MgCl2:10mM', 'chemical:ZMP:10uM', ...
'temperature:24C', ...
'modifier:SHAPE', ...
};
And the rest:
filename = 'pfl_2D_SHAPE_plus.rdat';
name = 'RNA Puzzle 13';
comments = { ...
'Preliminary data', ...
'RNA was heat to 90 C for 2 min and cool on ice for 2 min, then incubated at 37 C for 20 min at pH 8.0,', ...
' in presence of 10 uM ZMP ligand with 10 mM MgCl2 to aid folding.', ...
'Note: ', ...
'Taken in early May 2015 for thirteenth RNA Puzzle community-wide modeling trial.', ...
};
output_workspace_to_rdat_file(filename, name, sequence, offset, seqpos, area_peak, ...
structure, annotations, data_annotations, darea_peak, [], [], [], comments);
d_rdat = show_rdat(filename);