Most Useful Commands
-
rr(), orrender_rna()
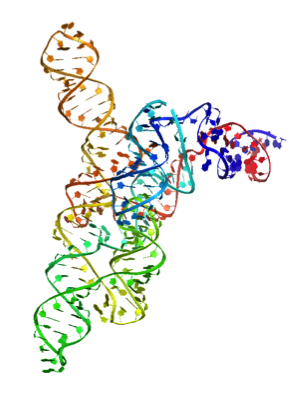
Rhiju’s favorite coloring of RNA with 2′ OH as spheres, bases as filled rings, and backbone as cartoon ribbons, rainbow colored from 5′ to 3′. No hydrogens, white background.
-
rrs(), orrender_rna_sticks()
Rhiju’s favorite coloring of RNA, showing all heavy atoms as sticks – more detail than
rr().
-
color_by_data( filename [, offset = 0, min = 0, max = 1] )
Read in a text file with rows and color specified residue numbers by scalar values.
125 0.12
126 1.50
Takes advantage of B-factor column and color by temperature function in pymol.
By default, coloring is scaled/offset based on lowest/highest scalar value. To specify the minimum and maximum values for coloring, enter a min and max value as shown above. For normalized chemical mapping reactivities,
min = 0andmax = 1~1.5usually works.
For instructions on coloring 3D models using analyzed reactivity data from HiTRACE, see instructions here.
-
rd(), orrender_molecules()
Rhiju’s favorite coloring of proteins and generic molecules side chains are all-heavy-atom and colored CPK, backbone is rainbow cartoon from N to C terminus.
-
loop_color( start, end, native=None, zoom=False )
Used for rendering protein loop modeling puzzles. White in background, colored red/blue over loop.
-
align_all( [ subset=[] ] )
Superimpose all open models onto the first one. This may not work well with selections.
Additional Commands
-
rc(), orrender_cartoon()
Tube coloring for large RNA comparisons
-
sa( [intra=False, rainbow=True] ), orsuperimpose_all()
Superimpose all open models onto the first one. This may not work well with selections.
Option
intracan be set toTruetoenable intra_fit()first, for working with multi-state (nmr) pdbs. [Thanks to Kyle Beauchamp for this one]
-
chainbow()
Run chainbow on all molecules, one by one.
-
rx(), orrender_x()
Rhiju’s favorite coloring of proteins, more details – no cartoon; heavy backbone. Use for beta peptide visualization.
-
rj(), orrender_rhiju()
Rhiju’s residue-level favorite coloring of proteins non-polar side chains colored gray.
-
rr2(),render_rna2()
Rhiju’s favorite coloring of RNA, showing all heavy atoms as sticks – more detail than
rr().
-
get_residue_colors( sele )
Get RGB color values associated with a selection. Useful if you want to exactly match coloring of 3D models with coloring in, say, a MATLAB script.
-
spr(), orsource_pymol_rhiju()
Load up these commands again after, say, an edit.
-
rb()
Basic cartoon coloring